TL;DR here is public gist
Introduction
I had a conversation with 9 about clustering links based on content and he pointed me to a StackOverflow(SO) conversation about getting started with it. It is very simple concept, but I had to stare at the pseudo code of paper, StackOverflow, results of different sample-sets, for quite some time to understand what is actually happening.
n = 0, t = false
v V do n = n + 1, c(v) = n end
while t = false do
t = true
v V do
c = i if (u, v) is max.
if c(v) != c then c(v) = c , t = false
end
end
For example dataset mentioned in StackOverflow
texts = [
"foo blub baz",
"foo bar baz",
"asdf bsdf csdf",
"foo bab blub",
"csdf hddf kjtz",
"123 456 890",
"321 890 456 foo",
"123 890 uiop",
]
If we try to plot initial graph based on cosine distance among all
the nodes(each element of texts list) we get something like this:
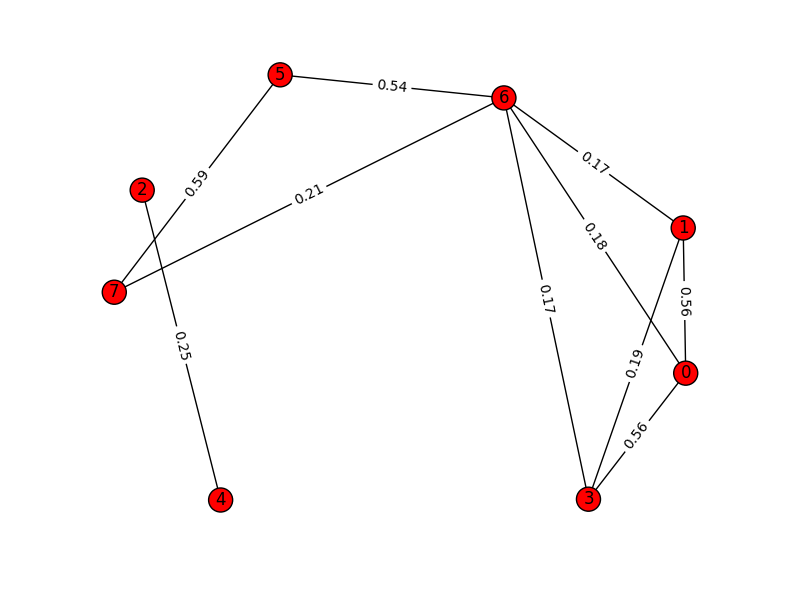 And once we cluster the graph using MajorClust algorithm, the graph
get restructured to:
And once we cluster the graph using MajorClust algorithm, the graph
get restructured to: 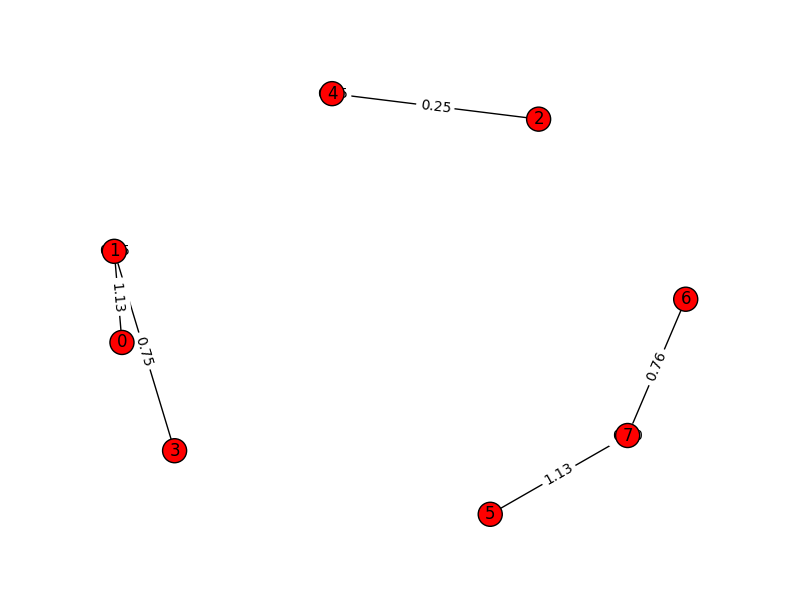
Notice how edges get aggregated and new weights are assigned to edges based on clusters and not-so-important edges are dropped altogether.
Code
The code on StackOverflow thread is complete without any external dependencies. I used TfidfVectorizer from sklearn to create vector space model from the documents(different and bigger dataset).
# corpus is list of all the documents
vectorizer = TfidfVectorizer(stop_words = "english",
ngram_range = (2, 3),
max_features = 100000)
corpus_mat = vectorizer.fit_transform(corpus)
num_of_samples, num_of_features = corpus_mat.shape
Then, I calculate cosine distance of each vector(document) from other vectors.
magnitudes = np.zeros((num_of_samples))
# this loop can be removed?
for doc_id in range(num_of_samples):
magnitudes[doc_id] = np.sqrt(corpus_mat[doc_id].dot(corpus_mat[doc_id].T).sum())
cosine_distances = np.zeros((num_of_samples, num_of_samples))
# this loop can be improved
for doc_id, other_id in combinations(range(num_of_samples), 2):
distance = (corpus_mat[doc_id].dot(corpus_mat[other_id].T).sum())/(magnitudes[doc_id]*magnitudes[other_id])
cosine_distances[doc_id, other_id] = cosine_distances[other_id, doc_id] = distance
This cosinedistances matrix is adjacency matrix representation of the graph, where each node is one document and edge weight(cosine distance) being the measure of similarity among two documents. Now MajorClust algorithm is clustering based on aggregating/accumulating edge weights and assigning each node to cluster which has maximum accumulated edge weight.
t = False
indices = np.arange(num_of_samples)
while not t:
t = True
for index in np.arange(num_of_samples):
# aggregating edge weights
new_index = np.argmax(np.bincount(indices,
weights=cosine_distances[index]))
if indices[new_index] != indices[index]:
indices[index] = indices[new_index]
t = False
clusters = {}
for item, index in enumerate(indices):
clusters.setdefault(index, []).append(links[-50+item]["url"])
This bit might be confusing, I was stuck here for quite some time, maybe images of graph above might help.
Experimentation and results
For testing the performance of algorithm I took facebook pages of three websites techcrunch, arstechnica, kafila(two of them cover articles related to tech, science and third one is about Indian politics and media). Using fbconsole I got last 50 links(only english stories) which have been shared, scraped content of those links and ran MajorClust over this data. I had to play around with Tf-Idf parameters like using stop words, ngrams(2 and 3) to get decent results. Once I had it, I ran it on single group(kafila) to create more clusters out of posted stories.
Next..
- Try to profile FB friend circle based on links they are sharing.
- Other clustering algorithms, k-means etc.
- Running this algorithm with different situations(like streaming data).